Molecules 2.0 now on the App Store
A major new update to Molecules, version 2.0, is now live on the App Store. This version brings an all-new renderer for newer iOS devices, one that is capable of beautiful, realistic 3-D graphics. You are now able to search the NCBI PubChem database of small molecule compounds, something people have been asking for since the launch of the application. In addition to this, the interface of the application has been streamlined and particularly improved on the iPad. As always, the latest source code of Molecules is available for download.
New rendering engine
The single biggest change in the application is the new rendering engine. For iOS devices that support OpenGL ES 2.0, this engine delivers stunning 3-D visualizations of molecular structures, unlike anything seen on a mobile device to date. The devices that can use this engine include the iPhone 3G S and anything newer (iPhone 4, third and fourth generation iPod touch, iPad, iPad 2, and whatever new things Apple will throw at us).
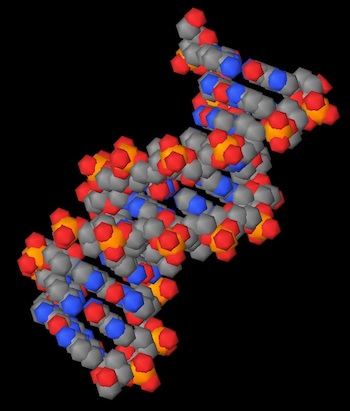
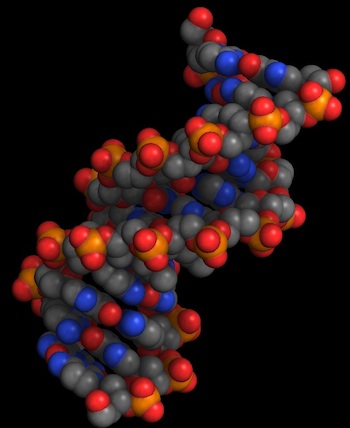
To give you an idea of the difference in quality, the following is a comparison of the old rendering engine (left) and the new one (right):
The new engine draws true smooth spheres and cylinders, as opposed to the polygonal approximations used in the previous versions of Molecules. This not only makes the spacefilling visualization mode a lot clearer and more useful, it also makes practical the display of small molecules (something enhanced by the PubChem support that I'll discuss later).
A more subtle enhancement of the 3-D display is the use of ambient occlusion shadowing in the molecular models. This means that recessed areas are darker than exposed ones, just like objects in the real world. This shadowing gives the models a more physical feel, but more importantly it can make clear the external structure of proteins and other large biomolecules by letting you see pockets and other elements on the surface of the molecule.
I'll have a more complete writeup of how I implemented this new style of rendering in a later post. For now, I can say that it is based on pioneering work by Marco Tarini, Paolo Cignoni, and Claudio Montani from the Universita dell’Insubria and I.S.T.I.-C.N.R, as demonstrated in their open source application QuteMol. While QuteMol was a great example of this technique, I chose to reimplement this from the ground up to be better integrated with iOS and the capabilities of mobile GPUs.
I should thank Jeff LaMarche for the publication of his GLProgram helper class for managing shaders, which he posted as part of the chapters from his unwritten OpenGL ES 2.0 book. I used this class to simplify the use of shader programs within this version of the application.
Because of the nature of the rendering in this new version, I needed to use language features that only exist in iOS 4.0. Unfortunately, this means that I cannot support iPhone OS 3.x any more, so Molecules will no longer run on this OS version. I'm sorry for those with original iPhones and iPod touches that cannot run iOS 4.0, but I was going to have to drop iPhone OS 3.x support at some point. As long as you don't update the application within iTunes, you should be able to keep using the old version on these devices.
PubChem database support
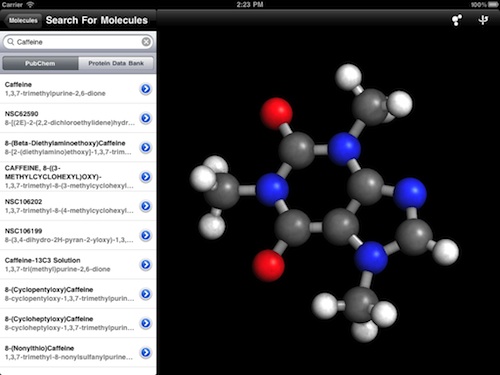
Now that the new rendering engine allows for slick representations of even small molecules, I've added a second database source for molecular structures on top of the existing one for the RCSB Protein Data Bank: NCBI's PubChem. PubChem is a massive public database of over 32 million chemical compounds, and is part of the NIH's Molecular Libraries Roadmap Initiative.
You can perform keyword searches against this database and download the small molecule structures contained therein. This should be a tremendous help to chemistry students and teachers, because the large biomolecules of the Protein Data Bank might be harder for them to understand than compounds like acetone and benzene.
I'd like to personally thank Dr. Rana Morris and the developers at PubChem for helping to make this feature possible.
Other enhancements
Beyond the major additions, I've streamlined the interface on both iPhone and iPad, although the iPad interface got the most love in this version.
A few bonds were being rendered incorrectly in a couple of residues, so those have been fixed.
I've also added a bunch of elements that weren't being recognized previously, which should now cover all of the common elements in both the Protein Data Bank and PubChem structures.
Overall, I'm pretty excited about this version. I've wanted to implement rendering like this within Molecules for years, but didn't understand the fundamentals well enough to actually make it work. The time I spent with OpenGL ES 2.0 for my Advanced iOS Development course and the talk I gave at SecondConf gave me the confidence to finally give this a try. I couldn't be happier with the results.